Feature Extraction¶
The first stage in consists in transforming the raw data into a uniform data matrix which will subsequently be given as input to the learning algorithm.
The current implementation of actsnclass text-like data from the SuperNova Photometric Classification Challenge
(SNPCC) which is described in Kessler et al., 2010.
Processing 1 Light curve¶
The raw data looks like this:
SURVEY: DES
SNID: 848233
IAUC: UNKNOWN
PHOTOMETRY_VERSION: DES
SNTYPE: 22
FILTERS: griz
RA: 36.750000 deg
DECL: -4.500000 deg
MAGTYPE: LOG10
MAGREF: AB
FAKE: 2 (=> simulated LC with snlc_sim.exe)
MWEBV: 0.0283 MW E(B-V)
REDSHIFT_HELIO: 0.50369 +- 0.00500 (Helio, z_best)
REDSHIFT_FINAL: 0.50369 +- 0.00500 (CMB)
REDSHIFT_SPEC: 0.50369 +- 0.00500
REDSHIFT_STATUS: OK
HOST_GALAXY_GALID: 17173
HOST_GALAXY_PHOTO-Z: 0.4873 +- 0.0318
SIM_MODEL: NONIA 10 (name index)
SIM_NON1a: 30 (non1a index)
SIM_COMMENT: SN Type = II , MODEL = SDSS-017564
SIM_LIBID: 2
SIM_REDSHIFT: 0.5029
SIM_HOSTLIB_TRUEZ: 0.5000 (actual Z of hostlib)
SIM_HOSTLIB_GALID: 17173
SIM_DLMU: 42.276020 mag [ -5*log10(10pc/dL) ]
SIM_RA: 36.750000 deg
SIM_DECL: -4.500000 deg
SIM_MWEBV: 0.0256 (MilkyWay E(B-V))
SIM_PEAKMAG: 22.48 22.87 22.70 22.82 (griz obs)
SIM_EXPOSURE: 1.0 1.0 1.0 1.0 (griz obs)
SIM_PEAKMJD: 56251.609375 days
SIM_SALT2x0: 1.229e-17
SIM_MAGDIM: 0.000
SIM_SEARCHEFF_MASK: 3 (bits 1,2=> found by software,humans)
SIM_SEARCHEFF: 1.0000 (spectro-search efficiency (ignores pipelines))
SIM_TRESTMIN: -38.24 days
SIM_TRESTMAX: 64.80 days
SIM_RISETIME_SHIFT: 0.0 days
SIM_FALLTIME_SHIFT: 0.0 days
SEARCH_PEAKMJD: 56250.734
# ============================================
# TERSE LIGHT CURVE OUTPUT:
#
NOBS: 108
NVAR: 9
VARLIST: MJD FLT FIELD FLUXCAL FLUXCALERR SNR MAG MAGERR SIM_MAG
OBS: 56194.145 g NULL 7.600e+00 4.680e+00 1.62 99.000 5.000 98.926
OBS: 56194.156 r NULL 3.875e+00 2.752e+00 1.41 99.000 5.000 98.953
OBS: 56194.172 i NULL 3.585e+00 4.628e+00 0.77 99.000 5.000 99.033
OBS: 56194.188 z NULL -2.203e+00 4.463e+00 -0.49 99.000 5.000 98.983
OBS: 56207.188 g NULL -7.008e+00 4.367e+00 -1.60 99.000 5.000 98.926
OBS: 56207.195 r NULL -1.189e+00 3.459e+00 -0.34 99.000 5.000 98.953
OBS: 56207.203 i NULL 8.799e+00 6.249e+00 1.41 99.000 5.000 99.033
You can load this data using:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | >>> from actsnclass.fit_lightcurves import LightCurve >>> path_to_lc = 'data/SIMGEN_PUBLIC_DES/DES_SN848233.DAT' >>> lc = LightCurve() # create light curve instance >>> lc.load_snpcc_lc(path_to_lc) # read data >>> lc.photometry # check structure of photometry mjd band flux fluxerr SNR 0 56194.145 g 7.600 4.680 1.62 1 56194.156 r 3.875 2.752 1.41 ... ... ... ... ... ... 106 56348.008 z 70.690 6.706 10.54 107 56348.996 g 26.000 5.581 4.66 [108 rows x 5 columns] |
Once the data is loaded, you can fit each individual filter to the parametric function proposed by Bazin et al., 2009 in one specific filter.
1 2 3 | >>> rband_features = lc.fit_bazin('r') >>> print(rband_features) [159.25796385, -13.39398527, 55.16210333, 111.81204143, -20.13492354] |
The designation for each parameter are stored in:
It is possible to perform the fit in all filters at once and visualize the result using:
1 2 3 | >>> lc.fit_bazin_all() # perform Bazin fit in all filters >>> lc.plot_bazin_fit(save=True, show=True, output_file='plots/SN' + str(lc.id) + '.png') # save to file |
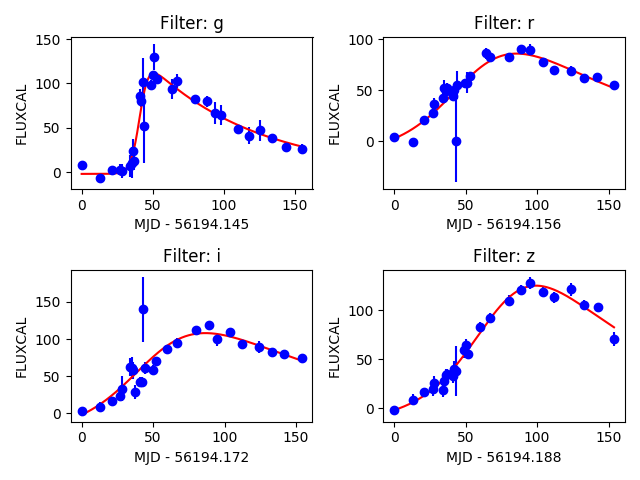
Processing all light curves in the data set¶
There are 2 way to perform the Bazin fits for the entire SNPCC data set. Using a python interpreter,
1 2 3 4 5 | >>> from actsnclass import fit_snpcc_bazin >>> path_to_data_dir = 'data/SIMGEN_PUBLIC_DES/' # raw data directory >>> output_file = 'results/Bazin.dat' # output file >>> fit_snpcc_bazin(path_to_data_dir=path_to_data_dir, features_file=output_file) |
The above will produce a file called Bazin.dat in the results directory.
The same result can be achieved using the command line:
>> fit_dataset.py -dd <path_to_data_dir> -o <output_file>